BY: RAHUL ANDHARIA (MSIWM001)
Nucleic Acid Hybridization: It is a technique which involves interaction of single stranded Nucleic acids(DNA/RNA) to form complexes called Hybrids, which contain the similar complementary sequences as that of nucleic acids. This technology helps us to understand the sequence identity among nucleic acids and to determine and detect specific sequences.
History:
In the year 1975, Grunstein and Hogness developed this method.
Principle:
In simpler terms it is a technique that simply detects specific DNA/RNA sequences using specific Probes. By using this technique we can determine sequence of hundreds of clones from a particular colony which exhibits it. Bacterial Colonies are transferred from agar plates to nitrocellulose membrane where they are lysed by using alkaline solution.
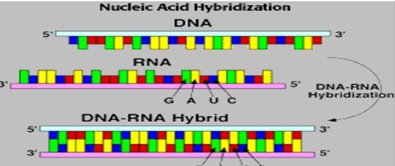
After cell lysis, due to the negative charge of nitrocellulose membrane, the DNA and proteins gets adhere to the membrane. To remove the bounded protein, the membrane is dipped in Proteinase k solution. Upon exposure to UV rays, the DNA gets fixed to the nitrocellulose membrane. After baking, the membrane gets exposed to labelled RNA for hybridization. The hybrids can be monitored by Autoradiography. Colony giving positive result must be picked from the master plate.
Procedure overview of Nucleic acid hybridization:
- Probe, (DNA/RNA fragment which can be fluorescently radiolabelled ) is required which will anneal to the target nucleic acid.
- The next step is the attachment of target to a solid matrix, like for example a membrane.
- Denaturation (breaking of hydrogen bonds) of the target and the probe used.
- The denatured probe is than added to a target in a solution (Proteinase k sol)
- If sequence homology exists between the probe and the target, the probe will than hybridize or will anneal the target.
- The probe which is hybridised than can be visualised by methods like Autoradiography, Chemiluminescence or Colorimetry.

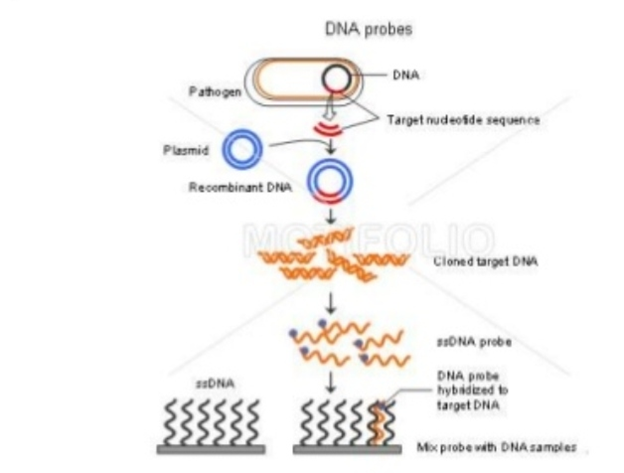
Hybridization Probes:
Probes are DNA or RNA fragments that can be radiolabelled. Probes help in identifying complementary segments in nucleic acid of Micro-organisms. The probe gets hybridised to single strands because of its complementarity with the target.
To make the probe hybridise with target sequence, it is labelled with molecular marker like radioactive 32-P ( radioactive isotope of phosphorus which incorporates into phosphodiester bonds of DNA).
Types of Probes:
- DNA Probe: Single stranded DNA is used to detect the complementary sequence amongother ss molecules. Therefore, it is a short sequence radiolabelled with radioactive isotope to detect complementary nucleotide sequence.
- RNA Probe: Single stranded RNA is used to detect the complimentary sequence among other ss molecules. RNA probes are also called as Riboprobes. RNA-RNA hybrids are known to be more stable than RNA-DNA hybrids generally because RNA hybrids shows more significant kinetic hydration.
- Oligonucleotide probes: refers to a short sequence of nucleotides which are synthesized to mimic the regions where mutations have occurred. They are synthesized in laboratory. Initially, a mononucleotide is attached to a solid support, and than one by one other mononucleotide are added to the 3’ end region. They are labelled with molecular markers at the 5’end.
Labelling of Hybridization Probes( DNA/RNA)
Involves 2 different types of methods:
- In vivo labelling: labelling of nucleotides in cultured cells.
- In vitro labelling: use of an enzyme to label the probe.
Strand synthesis labelling for DNA probes:
Most common method used to label DNA. In this method, enzyme DNA polymerase is used to attach labelled nucleotides to the DNA copies in the starting DNA. Any one of the nucleotide is labelled. There are various methods for DNA labelling:
- Nick Translation labelling: method in which DNA fragment is treated with DNase enzyme to induce single stranded nicks. Labelled nucleotides from nicked sites are incorporated by DNA polymerase I . Nick sites are replaced by DNA polymerase I that elongates 3’hydroxyl activity, thereby removing nucleotides by 5’-3’ exonuclease activity and replacing it with dntps.

- PCR Method: PCR method of amplification of DNA allows the resulting product to label either with modified nucleotide or oligonucleotide primers. With this, we can thus incorporate labelled nucleotides in the PCR reaction mixture which will than result in production of labelled PCR products throughout its length. This method has higher yield even with a little template.
Strand synthesis labelling of RNA probes:
Most common method used to label RNA. In this method, enzyme RNA polymerase is used to attach labelled nucleotides to the RNA copies in the starting RNA. Any one of the nucleotide is labelled. There are various methods for RNA labelling:
- In vitro transcription from cloned DNA inserts: The RNA probe gets transcribed from a linear DNA template with the help of bacteriophage DNA dependant-RNA polymerase enzymes from bacteria like salmonella (sp6) and E.coli(T3 and T7). One major advantage is with this method the labelled probes are completely free of the vector sequences.

- 3’end labelling RNA: Short DNA template has to be designed to anneal 3’end of RNA, with a 5’overhang of 3’ TA-5′. The klenow portion( large fragment of protein) of DNA polymerase I can than extend 3’end of RNA by incorporating single 32P radio labelled dAtp.
Uses of Nucleic Acid hybridization:
- Testing frequency of relatedness between 2 DNA molecules:
Sample of DNA 1 is heated to melt it to single stranded DNA. The ssDNA is than attached to a appropriate filter. Now chemical is being added to filter to block the sites, that can bind to DNA. After melting, DNA 2 sample is poured. Now some molecules from sample 1 will base pair with sample 2. The more closer the molecules are related, more hybrids it will generate. This method is highly useful, for example DNA of human gene, haemoglobin generated is melted and filtered, than it can be used to test the DNA from same gene of some different species.

- Isolating Genes for Cloning:
Suppose we have say human haemoglobin gene and want to isolate corresponding monkeys gene, than human DNA is added to filter and the monkeys DNA is cut into segments with the help of restriction enzymes. The monkey DNA is than heated to make it to single strand and poured in filter. The DNA fragment carrying monkeys haemoglobin gene will now bind with human haemoglobin gene and gets stuck in the filter. Other genes will not hybridize. This is a very selective approach and provides hybridization of new genes from related or similar kind of genes.
